Hello, I was wondering why the mutation step is used as mean in the Gaussian mutation sampled for CMA-ES. Doesn't that interfere with the use of the covariance to align the mutation with the gradient? (slide 19, lesson 2, Evolutionary Strategies)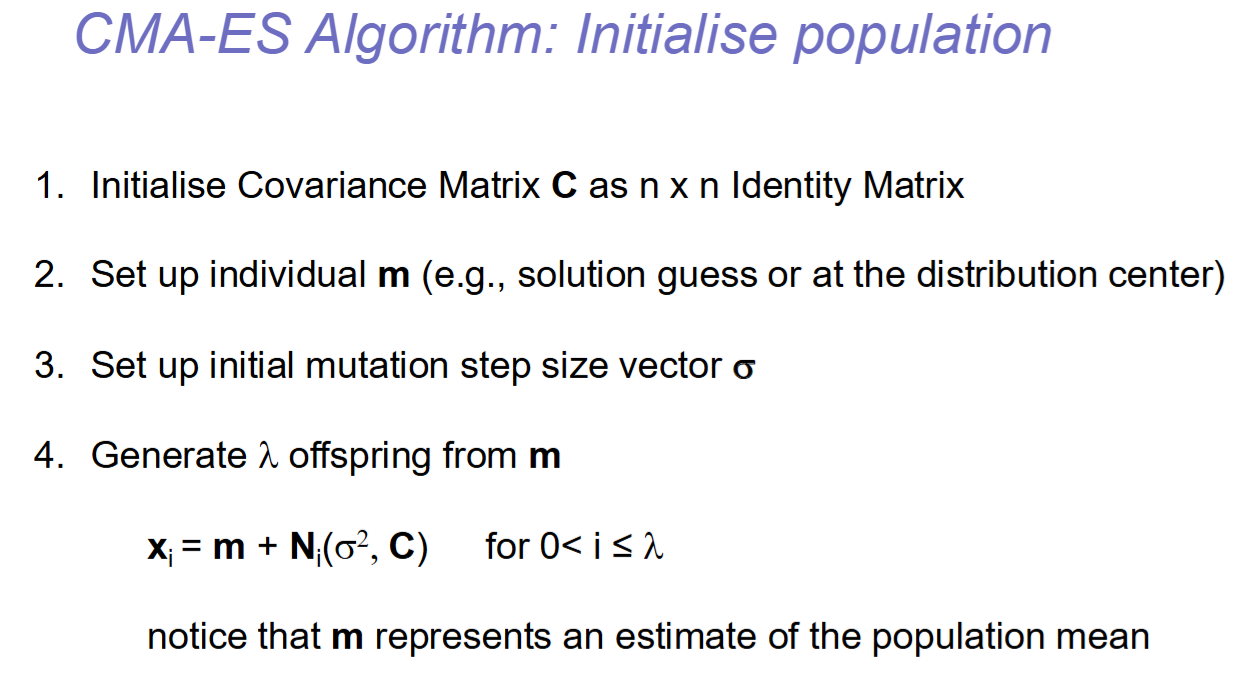
Hi,
There are different notations for CMAES that are quite similar.
A more conventional way of writing is:
x_i ~ N_i(m, sigma^2*C) , or,
x_i ~ m + sigma*N_i(0, C)
In essence, where the mutation ofsett is placed does not matter, e.g. m-sigma + N_i(sigma, C) is the same as the equation above.
You can assume any of these equations to be correct if it makes it more intuitive for you.
There are different notations for CMAES that are quite similar.
A more conventional way of writing is:
x_i ~ N_i(m, sigma^2*C) , or,
x_i ~ m + sigma*N_i(0, C)
In essence, where the mutation ofsett is placed does not matter, e.g. m-sigma + N_i(sigma, C) is the same as the equation above.
You can assume any of these equations to be correct if it makes it more intuitive for you.
